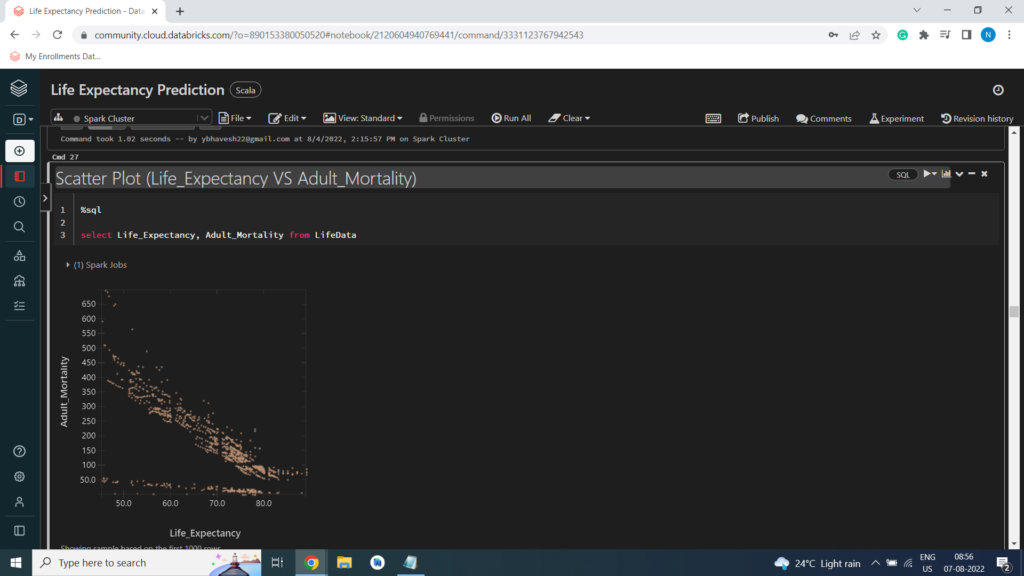
Scatter Plot (Life_Expectancy VS Adult_Mortality)
Scatter Plot (Life_Expectancy VS Infant_Deaths)

Scatter Plot (Life_Expectancy VS Alcohol)

Scatter Plot (Life_Expectancy VS Percentage_Expenditure)

Scatter Plot (Life_Expectancy VS Hepatitis_B)

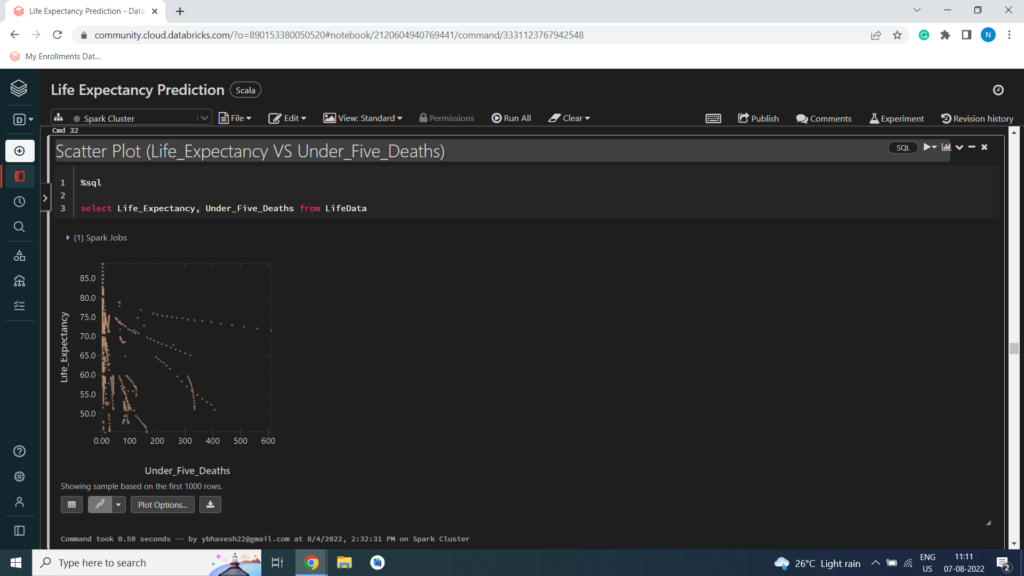
Scatter Plot (Life_Expectancy VS Under_Five_Deaths)
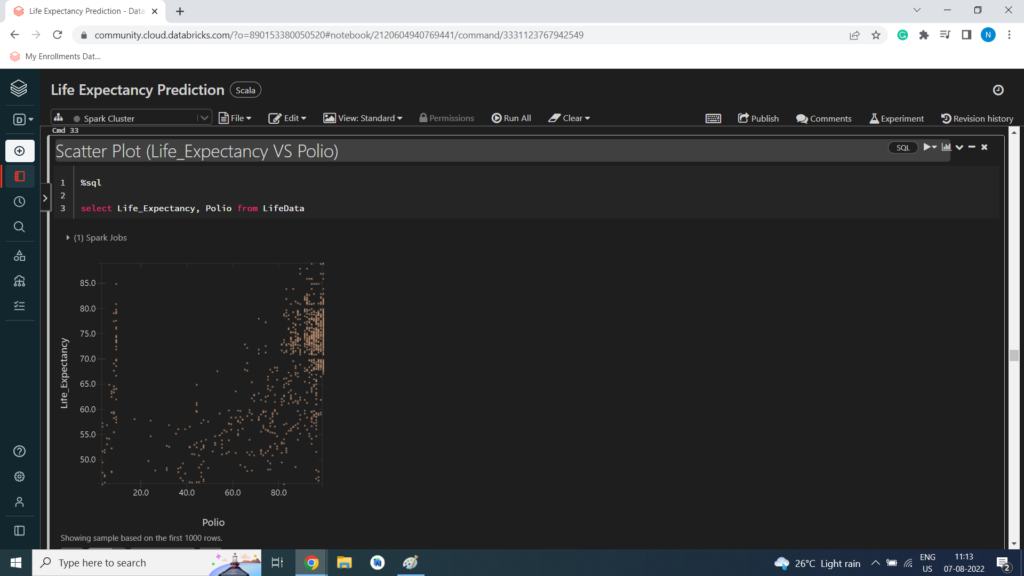
Scatter Plot (Life_Expectancy VS Polio)
Scatter Plot (Life_Expectancy VS Total_Expenditure)

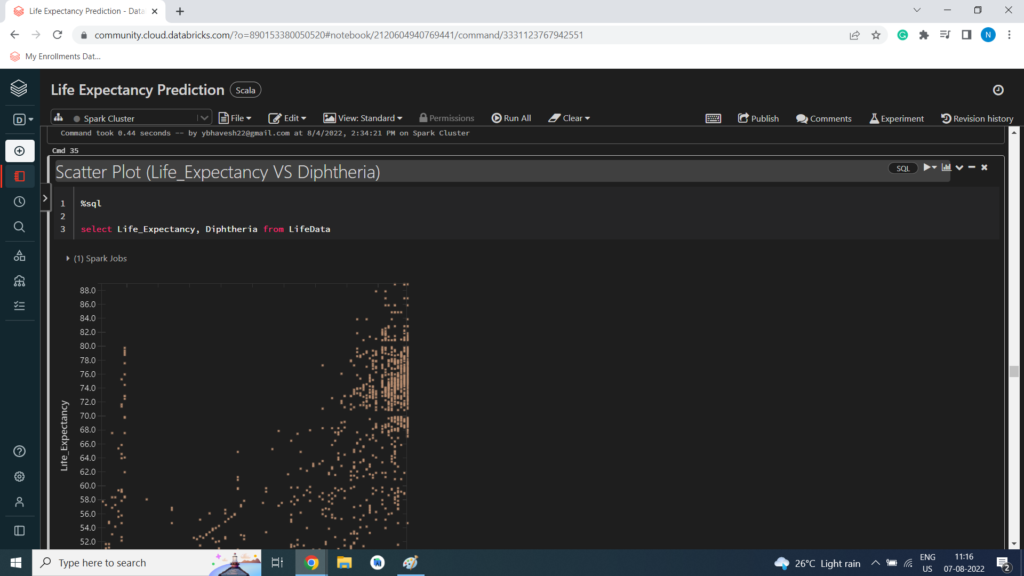
Scatter Plot (Life_Expectancy VS Diphtheria)
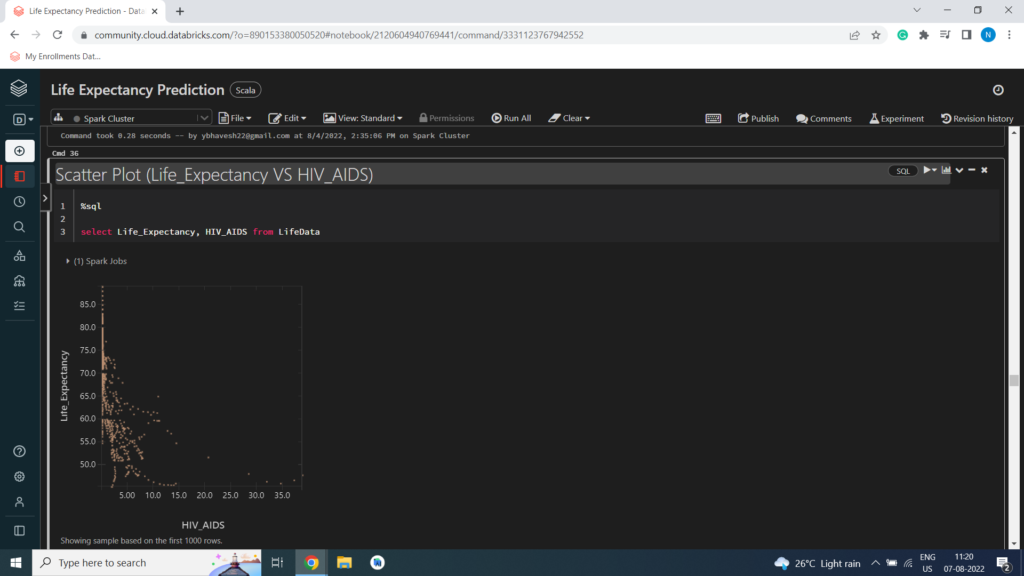
Scatter Plot (Life_Expectancy VS HIV_AIDS)
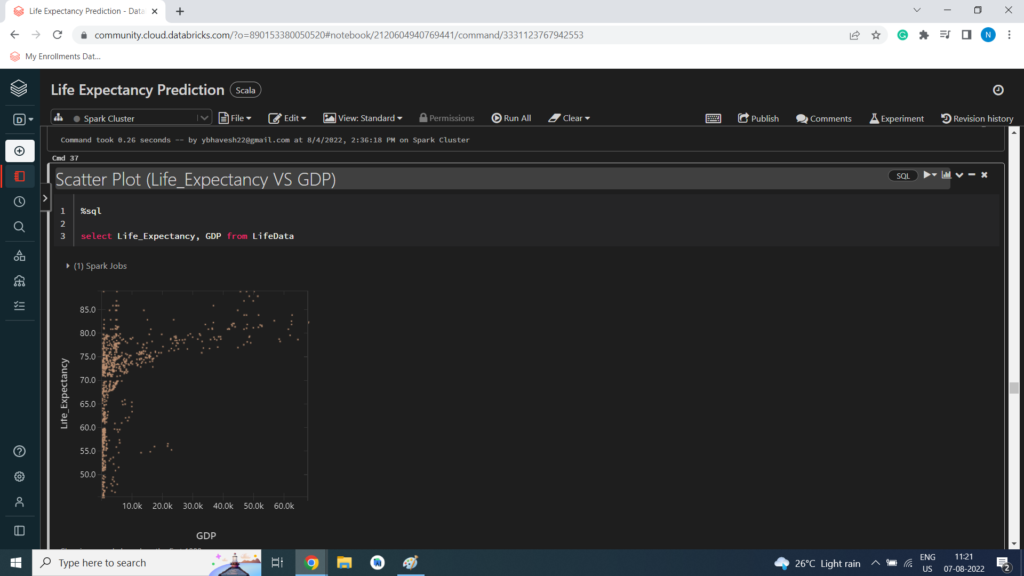
Scatter Plot (Life_Expectancy VS GDP)
Scatter Plot (Life_Expectancy VS Population)

Scatter Plot (Life_Expectancy VS Thinness_1_19_years)

Scatter Plot (Life_Expectancy VS Thinness_5_9_years)

Scatter Plot (Life_Expectancy VS Income_Composition_of_Resources)

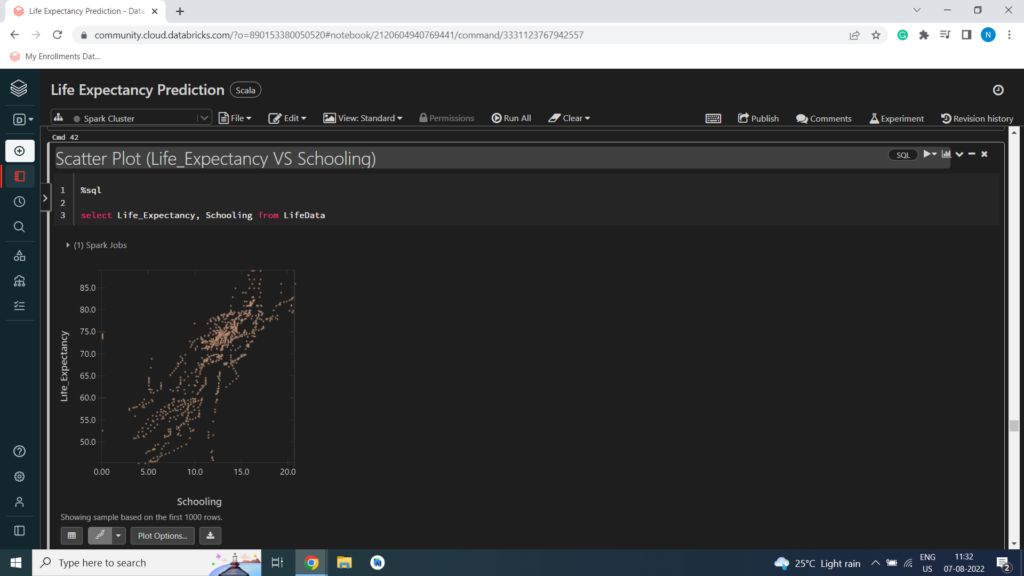
Scatter Plot (Life_Expectancy VS Schooling)
Scatter Plot (Schooling VS Adult_Mortality)

Scatter Plot (Schooling VS Income_Composition_of_Resources)

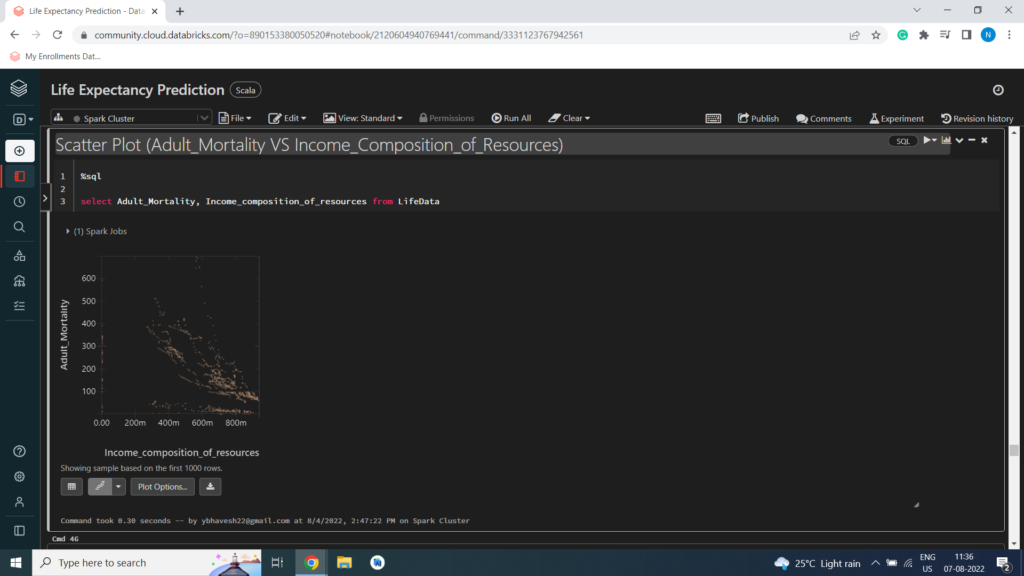
Scatter Plot (Adult_Mortality VS Income_Composition_of_Resources)
Collecting all String Columns into an Array
%scala
var StringfeatureCol = Array(“Country”, “Status”)
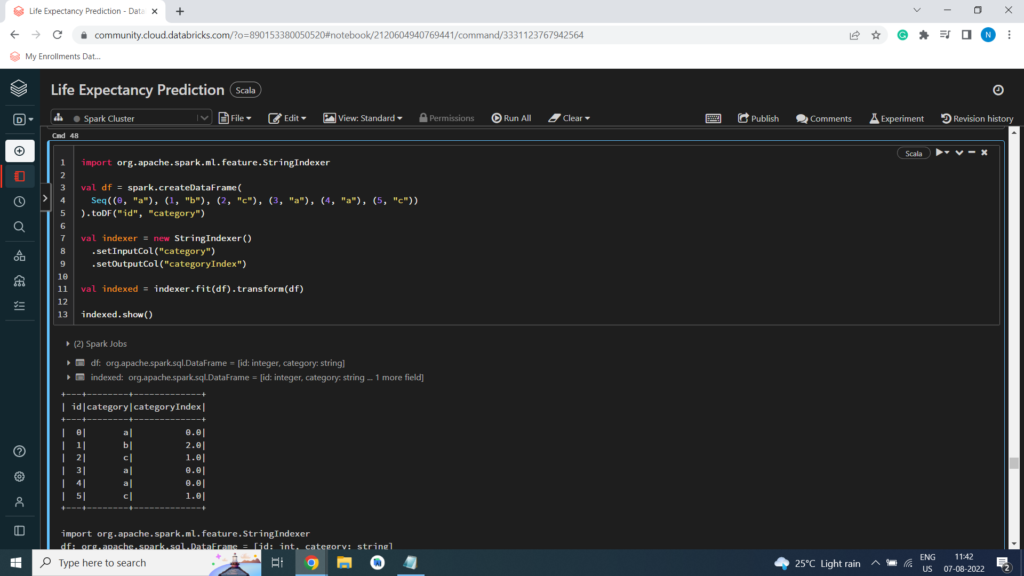
StringIndexer encodes a string column of labels to a column of label indices.
Example of StringIndexer
import org.apache.spark.ml.feature.StringIndexer
val df = spark.createDataFrame(
Seq((0, “a”), (1, “b”), (2, “c”), (3, “a”), (4, “a”), (5, “c”))
).toDF(“id”, “category”)
val indexer = new StringIndexer()
.setInputCol(“category”)
.setOutputCol(“categoryIndex”)
val indexed = indexer.fit(df).transform(df)
indexed.show()
Define the Pipeline
Define the Pipeline
A predictive model often requires multiple stages of feature preparation.
A pipeline consists of a series of transformer and estimator stages that typically prepare a DataFrame for modeling and then train a predictive model.
In this case, you will create a pipeline with stages:
A StringIndexer estimator that converts string values to indexes for categorical features
A VectorAssembler that combines categorical features into a single vector
%scala
import org.apache.spark.ml.attribute.Attribute
import org.apache.spark.ml.feature.{IndexToString, StringIndexer}
import org.apache.spark.ml.{Pipeline, PipelineModel}
val indexers = StringfeatureCol.map { colName =>
new StringIndexer().setInputCol(colName).setHandleInvalid(“skip”).setOutputCol(colName + “_indexed”)
}
val pipeline = new Pipeline()
.setStages(indexers)
val LifeDF = pipeline.fit(lifeDF).transform(lifeDF)
Print Schema
%scala
LifeDF.printSchema()
root
|– Country: string (nullable = true)
|– Year: integer (nullable = true)
|– Status: string (nullable = true)
|– Life_expectancy: double (nullable = true)
|– Adult_Mortality: integer (nullable = true)
|– infant_deaths: integer (nullable = true)
|– Alcohol: double (nullable = true)
|– percentage_expenditure: double (nullable = true)
|– Hepatitis_B: integer (nullable = true)
|– Measles: integer (nullable = true)
|– BMI: double (nullable = true)
|– under_five_deaths: integer (nullable = true)
|– Polio: integer (nullable = true)
|– Total_expenditure: double (nullable = true)
|– Diphtheria: integer (nullable = true)
|– HIV_AIDS: double (nullable = true)
|– GDP: double (nullable = true)
|– Population: double (nullable = true)
|– thinness_1_19_years: double (nullable = true)
|– thinness_5_9_years: double (nullable = true)
|– Income_composition_of_resources: double (nullable = true)
|– Schooling: double (nullable = true)
|– Country_indexed: double (nullable = false)
|– Status_indexed: double (nullable = false)
Split the Data
Split the Data
Split the Data
It is common practice when building machine learning models to split the source data, using some of it to train the model and reserving some to test the trained model. In this project, you will use 70% of the data for training, and reserve 30% for testing
%scala
val splits = LifeDF.randomSplit(Array(0.7, 0.3))
val train = splits(0)
val test = splits(1)
val train_rows = train.count()
val test_rows = test.count()
println(“Training Rows: ” + train_rows + ” Testing Rows: ” + test_rows)
Prepare the Training Data
To train the Linear Regression model, you need a training data set that includes a vector of numeric features, and a label column. In this project, you will use the VectorAssembler class to transform the feature columns into a vector, and then rename the Life Expectancy column to the label.
VectorAssembler()
VectorAssembler(): is a transformer that combines a given list of columns into a single vector column. It is useful for combining raw features and features generated by different feature transformers into a single feature vector, in order to train ML models like logistic regression and decision trees.
VectorAssembler accepts the following input column types: all numeric types, boolean type, and vector type.
In each row, the values of the input columns will be concatenated into a vector in the specified order.
%scala
import org.apache.spark.ml.feature.VectorAssembler
val assembler = new VectorAssembler().setInputCols(Array(“Country_indexed”, “Year”, “Status_indexed”, “Adult_Mortality”, “infant_deaths”, “Alcohol”, “percentage_expenditure”, “Hepatitis_B”, “Measles”, “BMI”, “under_five_deaths”, “Polio”, “Total_expenditure”, “Diphtheria”, “HIV_AIDS”, “GDP”, “Population”, “thinness_1_19_years”, “thinness_5_9_years”, “Income_composition_of_resources”, “Schooling”)).setOutputCol(“features”).setHandleInvalid(“skip”)
val training = assembler.transform(train).select($”features”, $”Life_expectancy”.alias(“label”))
training.show()

Train a Linear Regression Model
Train a Linear Regression Model
Next, you need to train a Linear Regression model using the training data. To do this, create an instance of the Linear Regression algorithm you want to use and use its fit method to train a model based on the training DataFrame. In this project, you will use a Linear Regression algorithm – though you can use the same technique for any of the regression algorithms supported in the spark.ml API
%scala
import org.apache.spark.sql.types._
import org.apache.spark.sql.functions._
import org.apache.spark.ml.Pipeline
import org.apache.spark.ml.feature.VectorAssembler
import org.apache.spark.ml.regression.LinearRegression
val lr = new LinearRegression().setLabelCol(“label”).setFeaturesCol(“features”)
val model = lr.fit(training)
println(“Model Trained!”)
Prepare the Testing Data
Now that you have a trained model, you can test it using the testing data you reserved previously. First, you need to prepare the testing data in the same way as you did the training data by transforming the feature columns into a vector. This time you’ll rename the Life_expectancy column to trueLabel.
%scala
val testing = assembler.transform(test).select($”features”, $”Life_expectancy”.alias(“trueLabel”))
testing.show()
Test the Model
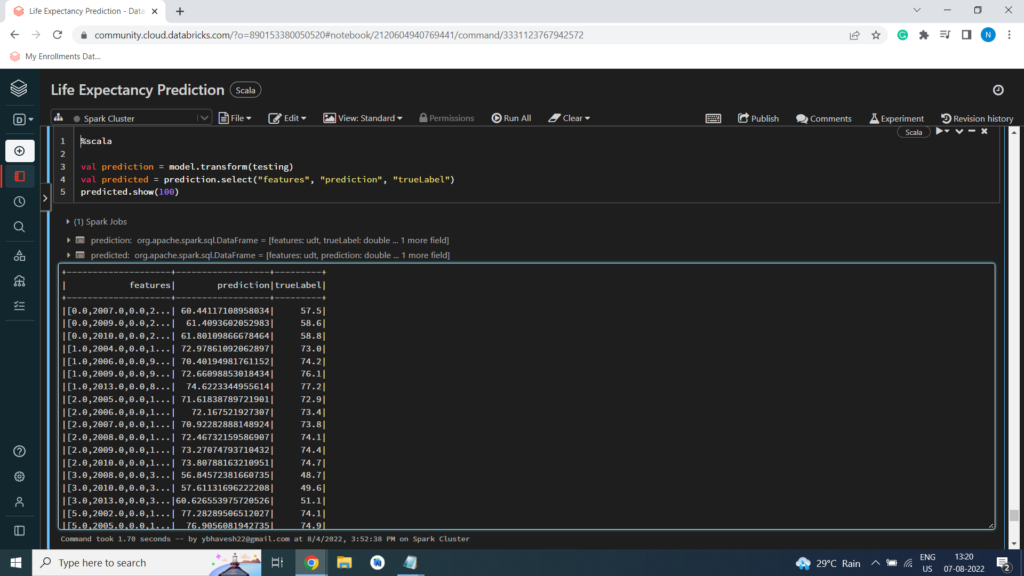
Now you’re ready to use the transform method of the model to generate some predictions. You can use this approach to predict the Life_expectancy; but in this case, you are using the test data which includes a known true label value, so you can compare the Life_expectancy
%scala
val prediction = model.transform(testing)
val predicted = prediction.select(“features”, “prediction”, “trueLabel”)
predicted.show(100)
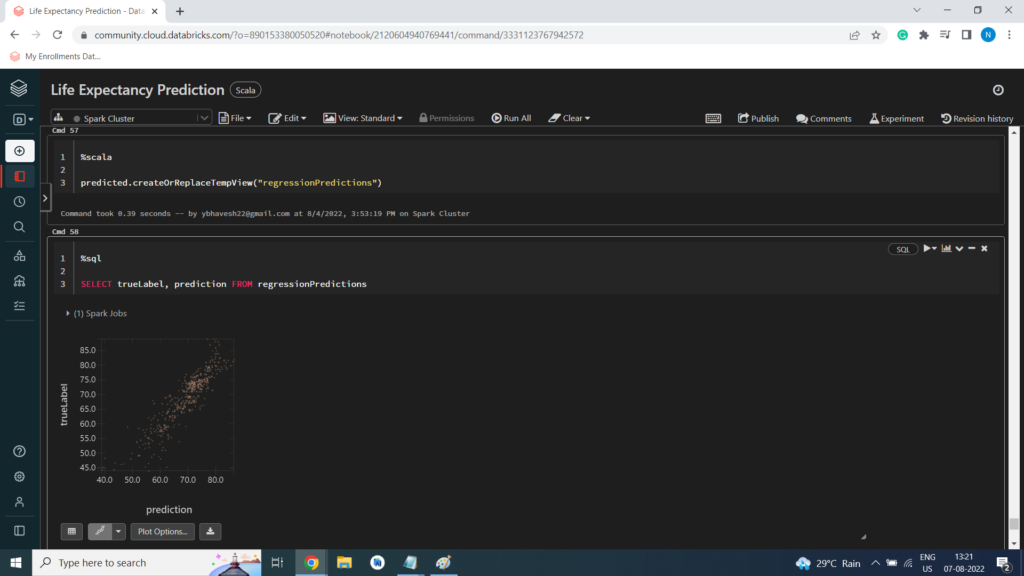
Root Mean Square Error (RMSE)
import org.apache.spark.ml.evaluation.RegressionEvaluator
val evaluator = new RegressionEvaluator().setLabelCol(“trueLabel”).setPredictionCol(“prediction”).setMetricName(“rmse”)
val rmse = evaluator.evaluate(prediction)
println(“Root Mean Square Error (RMSE): ” + (rmse))
Output:
Root Mean Square Error (RMSE): 3.775734673285438 import org.apache.spark.ml.evaluation.RegressionEvaluator evaluator: org.apache.spark.ml.evaluation.RegressionEvaluator = RegressionEvaluator: uid=regEval_5cd52eaaba66, metricName=rmse, throughOrigin=false rmse: Double = 3.775734673285438
